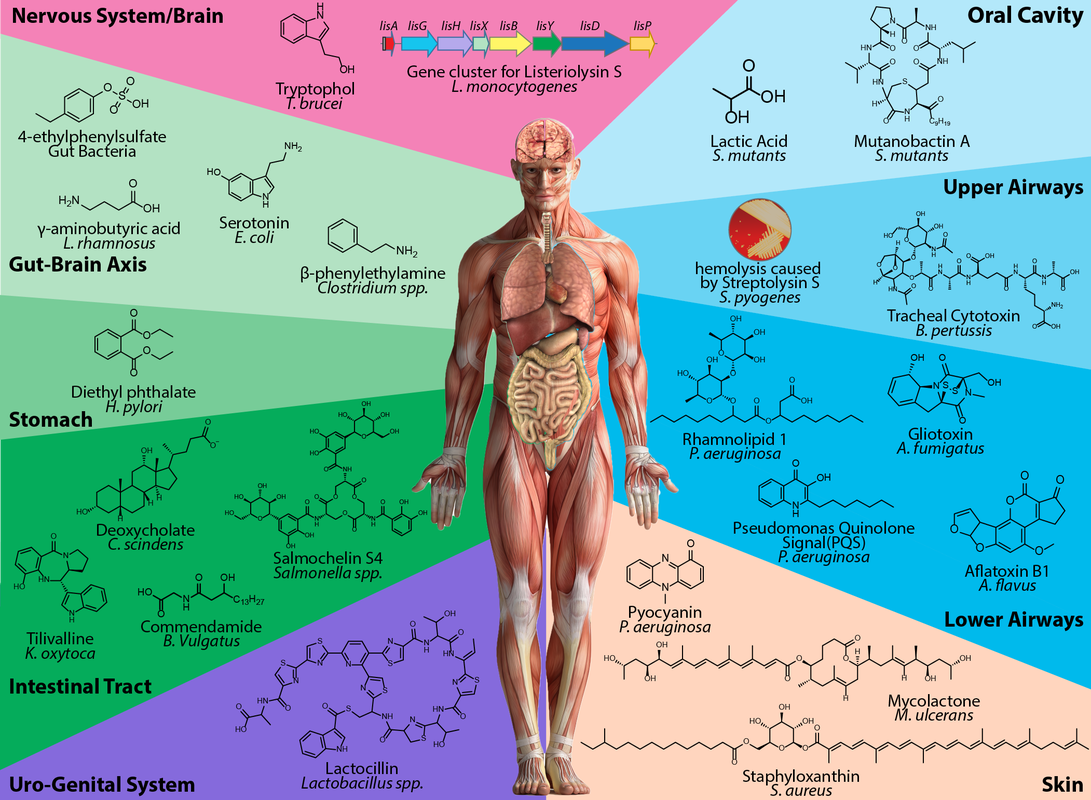
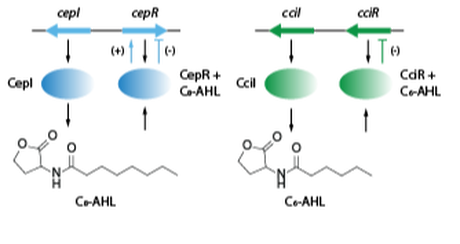
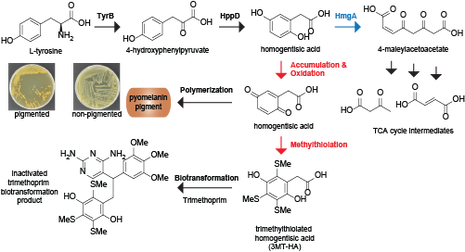
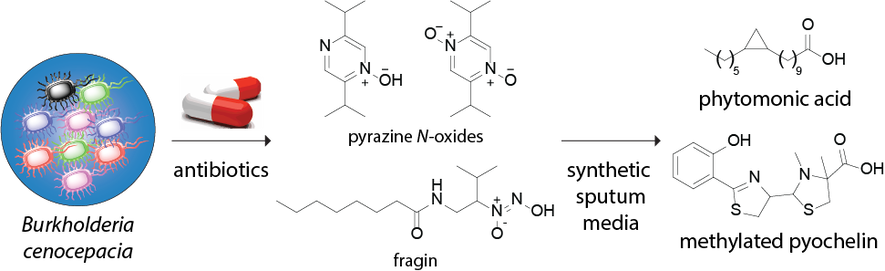
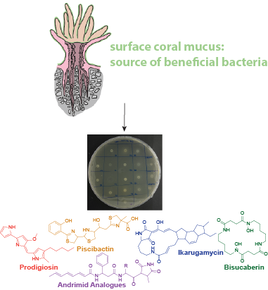
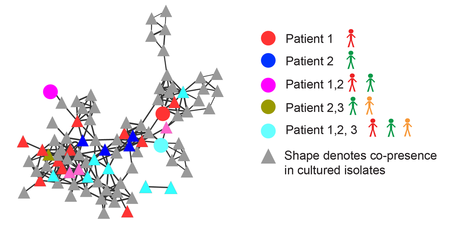
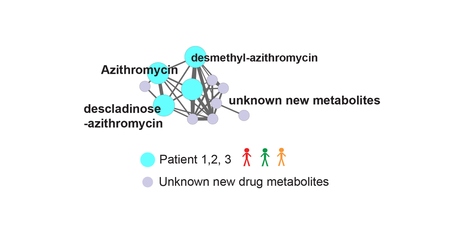
Microbial small molecules associated with host microbiome
Eukaryotes, including humans and marine organisms, are 'petri dishes', hosting an abundant and a rich prokaryotic 'microbiome'. The Garg Lab aims to understand the molecular interactions between a eukaryotic host and its microbiome, and how these molecular interactions affect health and disease status.
Using a variety of innovative tools including bioinformatics, microbiology, mass spectrometry, DNA sequencing, and 2D and 3D spatial mapping, we aim to delineate specific molecules that modulate the dynamics of microbial involvement in response to genetic and environmental triggers of disease.
Using a variety of innovative tools including bioinformatics, microbiology, mass spectrometry, DNA sequencing, and 2D and 3D spatial mapping, we aim to delineate specific molecules that modulate the dynamics of microbial involvement in response to genetic and environmental triggers of disease.
Biological and Chemical Interactions Spanning Different Scales
The Garg laboratory investigates natural product mediated chemical interactions at several scales
1. Phenotypic scale: Effect of microbial phenotypes such as quorum sensing and pigmentation on natural product chemical space of individual microbes
1. Phenotypic scale: Effect of microbial phenotypes such as quorum sensing and pigmentation on natural product chemical space of individual microbes
2. Microbe-Environment: Presence of antimicrobials and changes in nutrients
3. Microbe-Microbe
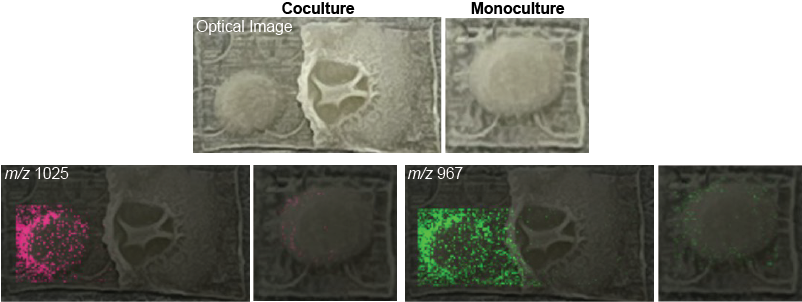
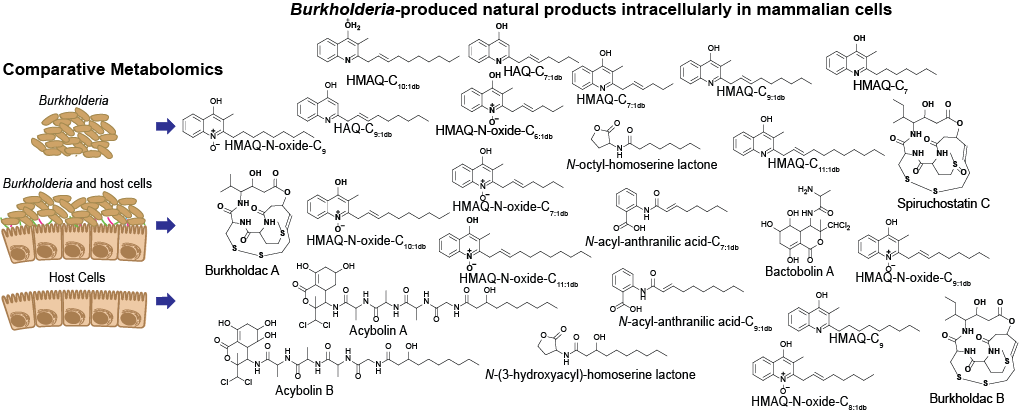
4. Microbe-host cell: Bacterial pathogen during infection of its host
5. Whole organism/holobiont
Tools used in our lab
The Garg laboratory uses a wide array of contemporary techniques in mass spectrometry complemented by (meta)genomics, microbiology, and biochemistry.
Mass spectrometry based three dimensional spatial mapping
We reconstruct 3-D models of human organs from CT scans and MRI images. We then overlay the spatial abundances of microbes (which we determine by DNA sequencing) and small molecules (which we find by mass spectrometry) on the human organ 3-D models to visualize correlations between the microbiome and the metabolome. These correlations allow us to determine (i) how the microbiome and the host are talking with each other at the molecular level using small molecules; (ii) whether the host is generating an immune response specific to the microbiome; (iii) whether clinically administered antibiotics and drugs affect the microbiome and the host; and (iv) how the microbiome-host community is structured. Our findings have broad implications on human health and disease and training opportunities exist for postdocs, graduate students, undergraduates, and research scientists interested in mass spectrometry, microbiology, and biochemistry.
|
3D organ mapping
|
Distribution of various antibiotics administered to the patients in the cystic fibrosis associated explant human lung.
|
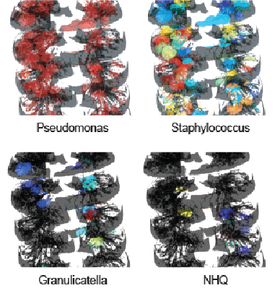
Distribution of the microbe Pseudomonas aeruginosa (left), Staphylococcus aureus (middle), and small molecule virulence factors produced by P. aeruginosa (right) in the cystic fibrosis associated human explanted lungs.
|
Mass spectrometry based molecular networking
The volume of information in a single mass spectrometry experiment can easily overwhelm manual curation and inventory. Then, how do we tackle a problem in which thousands of mass spectrometry experiments have to be conducted for a single dataset to tackle problems in human health and disease? The Garg lab uses techniques in molecular networking to visualize and organize large volumes of mass spectrometry data to derive meaningful correlations between metadata (conditions from which analytical samples were derived from) and the small molecule metabolome. These correlations are then tested and validated in the lab using a diversity of techniques in microbiology, molecular biology, and biochemistry. We also collaborate with clinicians to directly advance our findings to a clinical setting.
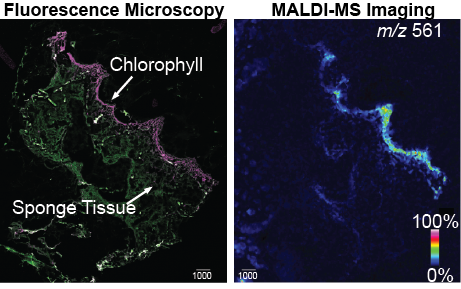
Mass spectrometry based imaging of polymicrobial communities
We use various wet lab techniques to mimic and understand molecular interactions in microbial communities. This allows us to test relevant hypotheses that are generated from direct analyses of human tissue using the abovementioned tools of spatial mapping and molecular networking. One of the tools that we use employs direct visualization of microbes interacting in a petri dish using mass spectrometry based microbial imaging. The Garg lab has access to cutting-edge mass spectrometers that allow high resolution imaging experiments to be performed with minimal sample manipulations. Our experiments are directed towards to questions that are directly relevant to human health, as well as complex community structures in the environment.
|
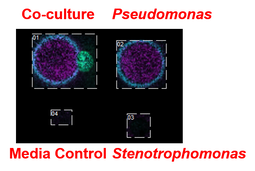
Stenotrophomonas specific small molecules produced only during interaction with Pseudomonas aeruginosa.
|
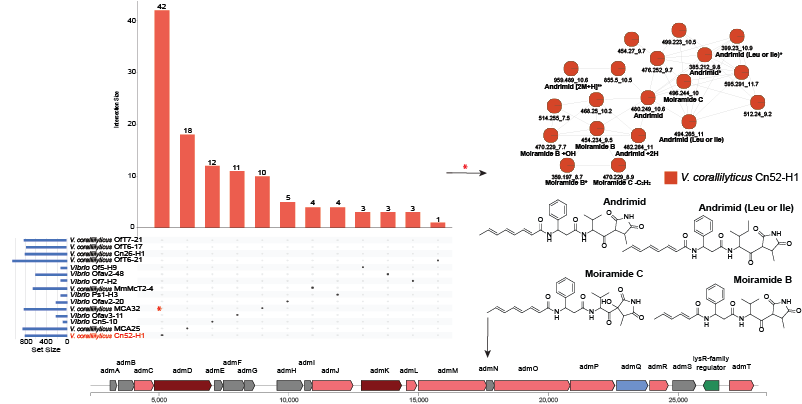
Genetic and biochemical bases for small molecule production
The microbial molecules that we find in complex communities using mass spectrometry are naturally produced using a set of dedicated genes encoded within the genomes of participating bacteria. We find these sets of genes, often organized in genetic neighborhoods called 'biosynthetic gene clusters', and use techniques in molecular biology, microbiology, and biochemistry to connect the genotype to the chemotype, and eventually to the phenotype in the community structure. We employ techniques involving expression of genes in different microbial hosts and biochemical characterization of biosynthetic enzymes.